Innate Immune Signaling in Health and Disease
The Yan laboratory studies homeostatic functions of innate immunity. We use inborn-error diseases of innate immunity as well as infection, cancer and neurodegenerative disease models to elucidate mechanisms of innate immune pathways and to develop novel therapies. We are particularly interested in nucleic acid immunity and type I interferon signaling. Current research includes:
1. The cGAS-cGAMP-STING DNA sensing pathway. We investigate mechanisms of STING trafficking and degradation by lysosomes, the role of STING in lysosome biology and lysosomal diseases, and dynamics of STING expression in tissues and immune cells. We also study DNase TREX1 that degrades cytosolic DNA to avoid cGAS-STING activation.
2. The OAS-2-5A-RNase L dsRNA sensing pathway. We study the physiological functions of OAS-2-5A-RNase L pathway in infection and cancer, including 2-5A as an immunotransmitter that spreads innate immunity. We also study the physiological functions of RNA exosome (degradation not secretion) in mammals, including maintaining skin tissue homeostasis and limiting the innate immune response to RNA via the OAS-RNase L pathway.
3. Neuroimmunology. We study the role of STING in neurodegenerative diseases, such as lysosomal storage disorders, NGLY1-deficiency, Parkinson's disease, Alzheimer's disease, amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD).
4. Cancer Immunology. We study the function of an innate immune checkpoint protein TREX1 and developing inhibitors. We study tumor-T cell interactions and mechanisms to enhance anti-tumor T cell immunity. We also study the role of immunotransmitters in cancer immunology.
Recent Publications:
Tang Z, Xing C, Araszkiewicz A, Yang K, Huai WW, Jeltema D, Dobbs N, Zhang Y, Sun LO, and Yan N. (2025) STING mediates lysosomal quality control and recovery through its proton channel function and TFEB activation in lysosomal storage disorders. Mol. Cell. 2025 Mar 27:S1097-2765(25)00201-1. PMID: 40185098
Knox K, Jeltema D, Dobbs N, Tang Z, Xing C, Song K, Torres-Ramirez G, Wu T, Yao C, and Yan N. (2025) Dynamic STING expression orchestrates immune cell development and function. Sci. Immunol 2025 Mar 7;10(105):eado9933. PMID: 40053603
Huai WW, Yang K, Xing C, Song K, Lyu H, Williams NS, Wu JJ, and Yan N. (2025) OAS Cross-Activates RNase L Intercellularly Through Cell-to-Cell Transfer of 2-5A to Spread Innate Immunity. Immunity. 2025 Feb 19:S1074-7613(25)00065-2. PMID: 40010341
Jeltema D, Knox K, Dobbs N, Siordia IR, Matthews J, Cohen M, Yan N. (2025) PARP7 is a negative feedback regulator of type I interferon signaling that protects against autoimmunity by inhibiting IRF3 transcriptional activity. J. Exp. Med. 2025 May 5;222(5):e20241184. PMID: 39969510
Our Research
STING Trafficking and Signaling Mechanisms
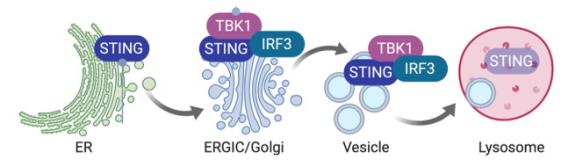
A unique feature of STING signaling is the obligatory trafficking of the STING protein through the secretory pathway. After ligand binding, STING undergoes a drastic conformational change that is believed to be the trigger for ER-exit. Then, STING translocates to the ER-Golgi intermediate compartment (ERGIC) and the Golgi, where it recruits kinase TBK1 and transcription factor IRF3. TBK1 phosphorylates itself, STING and IRF3. Phosphorylated IRF3 translocates to the nucleus and activates expression of IFN and IFN-stimulated genes (ISG). Although STING activation occurs on the Golgi, it does not dwell on the Golgi. Instead, STING rapidly moves pass the Golgi to the lysosome where it is degraded. ER-exit is a critical checkpoint for turning on STING signaling, and lysosomal degradation turns off STING signaling.
We showed that STING signaling can be activated by trafficking only, independently of ligand binding (Cell Host & Microbes 2015). We characterized STING gain-of-function SAVI mouse model and defined an IFN-independent function of STING (JEM 2017, 2019). We developed Sting-S365A mice to further define many other IFN-independent functions of STING (Immunity 2020). We uncovered cofactors of STING trafficking and signaling (PNAS 2017, Cell Report 2017, Nature Immunology 2020). We recently identified NPC1 as the lysosomal adaptor mediating STING protein degradation (Nature 2021).
Related publications:
- Dobbs N, Burnaevskiy N, Chen D, Gonugunta VK, Alto NM, Yan N (2015). STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host & Microbe. Aug 12;18(2):157-68. PMCID:PMC4537353
- Wu J, Chen YJ, Dobbs N, Sakai T, Liou J, Miner JJ, Yan N (2019). STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell death. J Exp Med. .216(4):867-883. PMID: 30886058.
- Pokatayev V, Yang K, Tu XT, Dobbs N, Wu JJ, Kalb RG and Yan N. (2020) Homeostatic regulation of STING protein at the resting-state by stabilizer TOLLIP. Nature Immunology. 21(2):158-167. PMID: 31932809
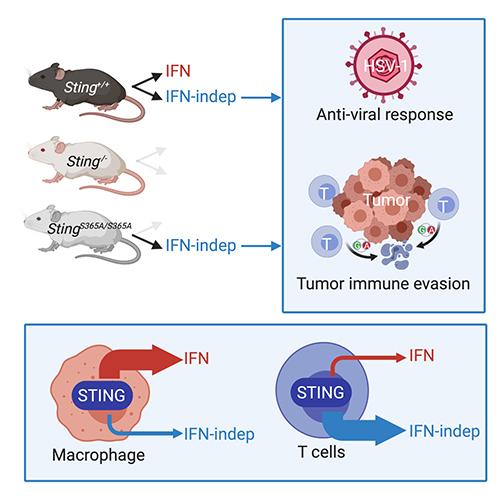
- Wu J, Dobbs N, Yang K and Yan N. (2020) IFN-independent activities of mammalian STING mediates antiviral response and tumor immune evasion. Immunity. Jul 14;53(1):115-126.e5. PMID: 32640258
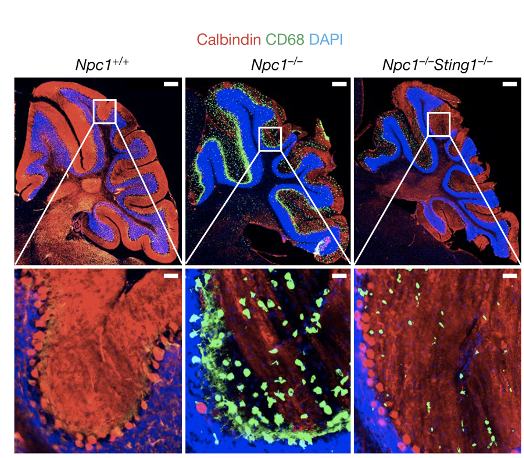
- Chu TT, Tu X, Yang K, Wu J, Repa JJ and Yan N. (2021) Tonic prime-boost of STING signaling mediates Niemann-Pick disease type C. Nature. 596, pages 570–575. PMID: 34290407
STING in Neurodegenerative Diseases
The STING pathway has been implicated in several neurodegenerative diseases, including Parkinson's disease (PD) and and amyotrophic lateral sclerosis (ALS). We recently found that tonic prime-boost of STING signaling mediates an early onset neurovegetative disease called Niemann-Pick disease type C (NPC). We also study another monogenic neurological disease in children called NGLY1-deficiency. We are interested in understanding how the STING pathway impacts functions of neuronal cells and whether we can target this pathway to treat neurodegenerative diseases.
- Yang K, Huang R, Suzuki T, Yan N (2018). N-glycanase NGLY1 regulates mitochondrial homeostasis and inflammation through NRF1.J Exp Med.Oct 1;215(10):2600-2616. PMID: 30135079
- Chu TT, Tu X, Yang K, Wu J, Repa JJ and Yan N. (2021) Tonic prime-boost of STING signaling mediates Niemann-Pick disease type C. Nature. 596, pages 570–575. PMID: 34290407
STING in Cancer Immunology
We are interested in understanding how innate immune signaling regulate cancer cell proliferation and tumorigenesis, and how can we harness innate immunity as novel cancer immunotherapy.
STING-mediated anti-tumor immunity through activating DC:T cell cross priming is well recognized. STING also has an anti-proliferation activity intrinsic to cancer cells. We recently found that STING activation in T cells causes T cell death and loss of tumor control. We are investigating whether tumor evades immune control by targeting the STING pathway in T cells.
- Gonugunta VK, Sakai T, Pokatayev V, Yang K, Wu JJ, Dobbs N, Yan N (2017). Trafficking-Mediated STING Degradation Requires Sorting to Acidified Endolysosomes and Can Be Targeted to Enhance Anti-tumor Response. Cell Reports. Dec 12;21(11):3234-3242.PMID: 29241549.
- Wu J, Dobbs N, Yang K and Yan N. (2020) IFN-independent activities of mammalian STING mediates antiviral response and tumor immune evasion. Immunity. Jul 14;53(1):115-126.e5. PMID: 32640258
Inborn Error Rare Disease
We are broadly interested in mechanisms of in inborn error immune diseases. We establish mouse models, investigate disease mechanism, then test potential therapies in mice.
In retinal vasculopathy with cerebral leukodystrophy (RVCL) associated with TREX1-frame—shift mutations, we identified the molecular defect in the glycotransferase OST enzyme complex (Immunity 2015), tested the FDA-approved drug ACM that inhibits the OST in mice in (J Autoimmunity 2017), and started a clinical trial using ACM to treat RVCL patients in 2017 (NCT02723448). We have since identified bioactive mammalian glycan species associated with autoimmune disease that are potent stimulators of innate immunity (Nat. Comm. 2019).
- Hasan M, Fermaintt CS, Gao N, Sakai T, Miyazaki T, Jiang S, Li QZ, Atkinson JP, Morse HC III, Lehrman MA and Yan N (2015). Cytosolic nuclease TREX1 regulates oligosaccharyltransferase activity independent of nuclease activity to suppress immune activation. Immunity.Sep 15;43(3):463-74.PMCID:PMC4575271
- Sakai T, Miyazaki T, Shin DM, Kim YS, Qi CF, Fariss R, Munasinghe J, Wang H, Kovalchuk AL, Kothari PH, Fermaintt CS, Atkinson JP, Perrino FW, Yan N*, Morse HC 3rd* (2017). DNase-active TREX1 frame-shift mutants induce serologic autoimmunity in mice.*Co-senior author. J Autoimmunity.Mar 18. pii: S0896-8411(17)30024-0. PMID:28325644. *Co-senior authors.
- Fermaintt CS, Sano K, Liu Z, Ishii N, Seino J, Dobbs N, Suzuki T, Fu YX, Lehrman MA, Matsuo I and Yan N.(2019). A bioactive mammalian disaccharide associated with autoimmunity activates STING-TBK1-dependent immune response. Nature Communications.10(1):2377.PMID: 31147550.
RNA Exosome Biology
The RNA exosome is an evolutionarily conserved intracellular RNA degradation machinery involved in RNA processing, maturation, surveillance and turnover. The super-killer (SKI) cytoplasmic RNA exosome is essential for survival in yeast but its physiological functions in mammals are unclear. Mutations in SKIV2L or TTC37 genes that encode key components of the SKI complex are associated with a rare inherited autosomal recessive disorder, trichohepatoenteric syndrome (THES). We recently showed that Skiv2l deficiency in mice disrupts skin epidermis and T-cell homeostasis. Skiv2l-deficient mice develop skin inflammation and hair abnormality that are also observed in a SKIV2L-deficient patient. Mechanistically, we demonstrate that mTORC1, a classical nutrient sensor, also senses cytoplasmic RNA quality control failure and drives autoinflammatory disease (Yang et al 2022 JCI). We also found that the RNA Exosome is important for early B cell development. Loss of function causes accumulation of ncRNA in the nucleus that impedes VDJ recombination during B cell development (Yang et al 2022 SI). More recently, we also show that RNA exosome limits innate immune response to RNA via the OAS-RNase L pathway (Yang et al 2024 EMBO J).
- Yang K, Han J, Asada M, Gill JG, Park JY, Sathe MN, Gattineni J, Wright T, Wysocki C, de la Morena MT, Garza LA, and Yan N. Cytoplasmic RNA quality control failure engages mTORC1-mediated autoinflammatory disease. J Clin Invest. 2022 Jan 18;132(2):e146176. PMID: 35040435
- Yang K, Han J, Gill JG, Park JY, Sathe MN, Gattineni J, Wright T, Wysocki C, de la Morena MT and Yan N. The Mammalian SKIV2L RNA exosome is essential for early B cell development. Science Immunology. 2022 Jun 3;7(72):eabn2888. PMID: 35658009
- Yang K, Dong B, Asthana A, Silverman RH, and Yan N. RNA helicase SKIV2L limits antiviral defense and autoinflammatory response through the OAS-RNase L pathway. EMBO Journal. 2024 Aug 7. PMID: 39112803