Approach
The McFadden lab uses a combination of patient-derived cancer models and genetically-engineered mouse models of cancer (GEMMs) to establish cancer cell line models suitable for high-throughput small molecule screens or genome-wide CRISPR screens. We collaborate with the UTSW High Throughput Screening (HTS) Core to perform small molecule screens, and work with medicinal chemists in the Department of Biochemistry to develop chemical probes for target identification studies. Our lab has also employed and expanded the use of forward genetics to identify compound-resistant alleles to small molecules with anti-cancer activity as an inroads to determination of molecular mechanism of action (Povedano et al., 2019 Cell Chemical Biology).
Thyroid cancer: We and others identified recurrent loss of function mutations in subunits of complex I of the electron transport chain (ETC) in Hürthle cell (oncocytic) carcinoma of the thyroid (Gopal, Kübler et al., Cancer Cell 2018, see reviews McFadden and Ganly, Thyroid 2019, and McFadden and Sadow, Frontiers in Endocrinology (2021). These mutations were interestingly encoded in the mitochondrial genome (mtDNA) rather than the nuclear chromosomal genome. Our lab is investigating the mechanisms underlying the selection for mutations that impair the function of the ETC, which is counterintuitive for cancers that should place a premium on metabolic adaptability.
Using genome wide CRISPR screening and patient-derived models of Hürthle cell carcinoma, our lab has recently shown using patient-derived models that mutations in the ETC in Hürthle cell carcinoma lead to a metabolic defect that can be targeted by inhibition of the lactate dehydrogenase (LDH) reaction. Because Hürthle cells are exclusively dependent on glycolysis for ATP generation, inhibition of LDH leads to an ATP crisis and rapid cell death in cell line and patient-derived xenograft models (Frank et al., Cancer Discovery, 2023; Frank et al., ACS Chemical Biology, 2024).
The lab continues to study the metabolic underpinnings of selection for ETC mutations and therapeutic approaches in thyroid cancer and in other forms of cancer including renal cell carcinoma.
Ewing Sarcoma: Ewing sarcoma is a lethal, debilitating cancer of children and young adults that is driven by a chromosomal translocation that leads to the formation of a neomorphic transcription factor, the EWSR1-FLI1 fusion oncoprotein. Like other cancers driven by fusion proteins, understanding the molecular mechanisms by which EWSR1-FLI1 drives oncogenesis represents an obvious tack to the development of effective targeted therapies for this disease.
The lab has generated patient-derived Ewing sarcoma (EWS) cell lines in which we have used CRISPR methods to knock in regulated protein degrons into the EWSR1-FLI1 chromosomal translocation. We have demonstrated an anti-proliferative, anti-tumor effect of EWSR1-FLI1 depletion in patient-derived cell lines and xenografts (McGinnis et al., 2024 bioRxiv.org). We then used the precise control of EWSR1-FLI1 protein levels using these degron systems to identify transcriptional readouts of EWSR1-FLI1 function. These readouts were then engineered with a series of nano-luciferase endogenous reporter alleles to perform a phenotypic HTS to identify small molecules that reproduce the genetic signature of EWSR1-FLI1 protein depletion. Our group is collaborating with UTSW Medicinal Chemists to identify the direct protein targets of small molecules that exhibit anti-EWS activity.
Small cell lung cancer:We have identified annotated (known or reported mechanism of action), and un-annotated small molecules from the UTSW HTS Core collection of small molecules (~350,000 individual chemical scaffolds) that suppress proliferation or survival of small cell lung cancer (SCLC) and other neuroendocrine cancer cell lines. These studies have led us to identify new covalent modifiers of microtubules (Povedano et al., 2020 J. Med Chem) and indirectly to the mechanism of action of a drug in clinical trials for Ewing sarcoma, TK216 (Povedano et al, 2022, Cell Chemical Biology). Our lab continues to study small molecules with activity against SCLC and other neuroendocrine cancers identified by this screening approach using medicinal chemistry and forward genetics.
Current and Past Funding
-
NIH Cancer Moonshot Program
-
Damon Runyon Cancer Research Foundation
-
St. Baldrick's Foundation
-
The Welch Foundation
-
1Million4Anna Foundation
Complex I mutations in thyroid cancer
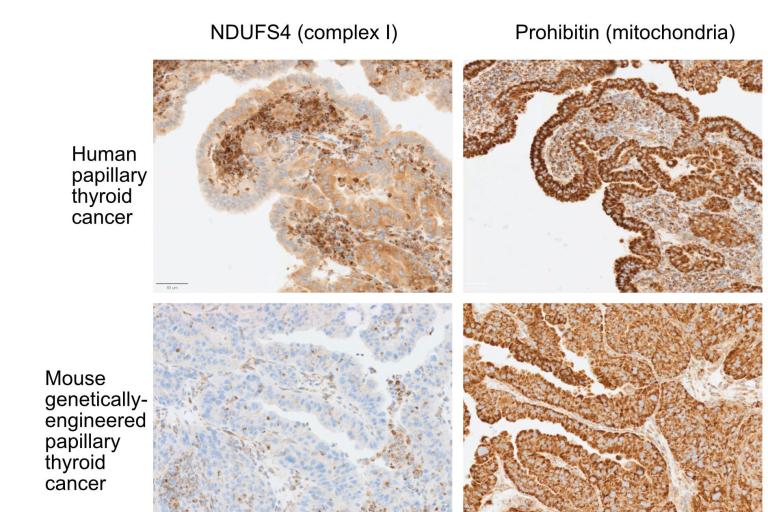 Loss of complex I in human and mouse thyroid cancer increases mitochondrial abundance
Loss of complex I in human and mouse thyroid cancer increases mitochondrial abundance Chemical screening
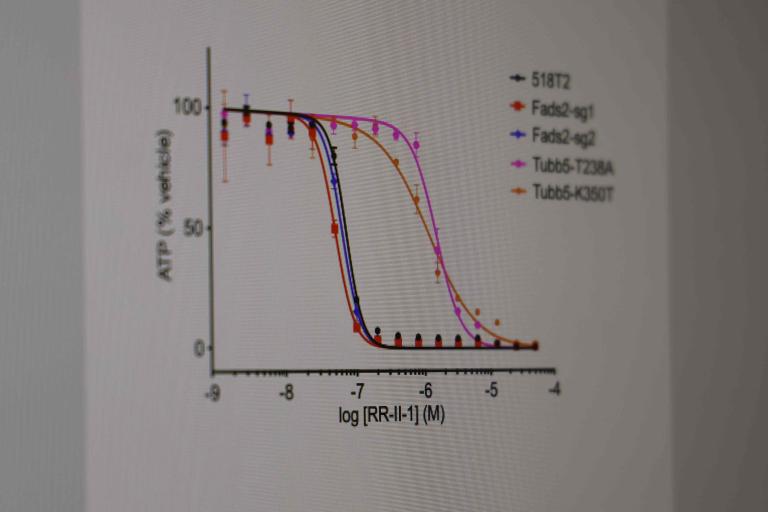 Identifying genotype- and lineage- selective cancer toxins.
Identifying genotype- and lineage- selective cancer toxins.