Organelle contact sites can spatially regulate Lipid Droplet dynamics
We are interested in how cellular organelles share information and metabolites such as lipids. This is of particular biomedical relevance, as recent work suggests intracellular trafficking of sterols and other dietary lipids is carefully regulated at sites of inter-organelle contact. As such, disruptions in lipid trafficking contribute to an array of genetic and acquired metabolic diseases (eg. Niemann Pick Type C, obesity, and heart disease).
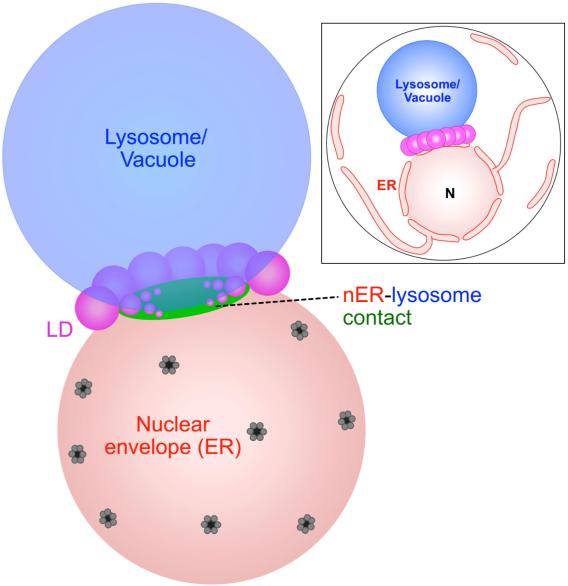
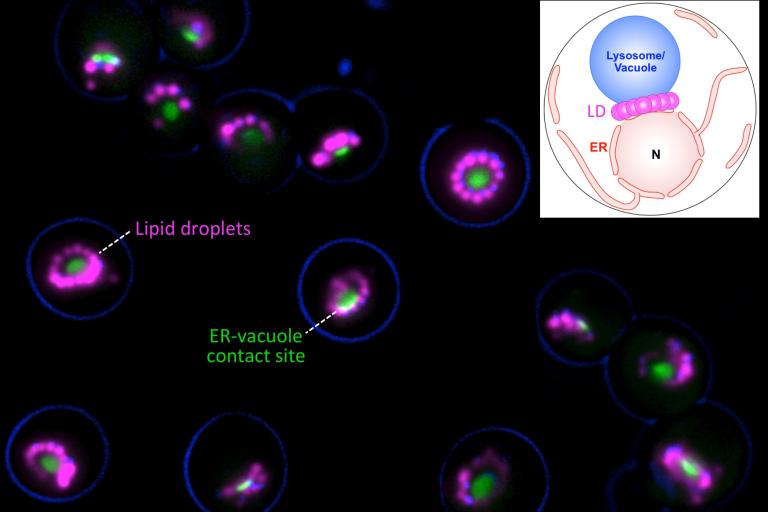
Recently, we made the unexpected discovery that organelle contact sites can spatially regulate the production and turn-over of lipid droplets (LDs), the primary energy-storing organelle of cells (Hariri, EMBO reports, 2018; Hariri, JCB, 2019). LDs store cellular fat as triglycerides (TAG, or TG), which can be mobilized during energetic crises like starvation or cell growth. We found that in yeast, LD biogenesis is spatially coordinated at the junction between the endosplasmic reticulum (ER) where LDs are made, and the vacuole/lysosome where LDs can be digested via a process called lipophagy. This ER-vacuole contact site is called the nucleus (nuclear ER)-vacuole junction (NVJ), and serves as a platform to organize lipid metabolism (see review Henne, COCB, 2019).
PXA domain-containing proteins organize LD biogenesis and LD spatial positioning
How do yeast cells spatially organize their LDs? Recently, we identified a new protein family that plays an important role in ER-endolysosomal cross-talk and LD biogenesis in yeast (Henne et al, JCB, 2015; Hariri, et al, EMBO reports, 2018). The yeast ortholog is Mdm1, which acts as an ER-vacuole tether but also interacts with LDs at ER-LD-vacuole tri-organelle contacts. Mdm1 and its orthologs all feature a poorly characterized PX-Associated (PXA domain), which binds lipids. Mdm1 and its mammalian and Drosophila homologs all appear to play roles in LD production and LD positioning in the cytoplasm by binding to LDs and attaching them to other cellular organelles (Hariri, et al, JCB, 2019; Datta, et al, JCB, 2019; Bryant, et al, HMG, 2018; Ugrankar, et al, Developmental Cell, 2019; Datta, PNAS, 2020). We also study how LD biogenesis and inter-organelle contacts relate to one another and influence cellular decision-making (Wood, Cell Reports, 2020).
Beyond their roles as inter-organelle tethers, PXA domain-containing proteins also function to promote LD biogenesis by recruiting and regulating fatty acid processing enzymes at ER sub-domains that produce LDs. Our lab is currently focused on understanding how these proteins promote LD biogenesis, and how the intracellular positioning of LDs promotes cellular homeostasis and adaptations to stress.